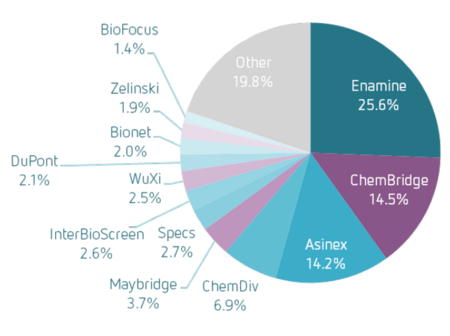
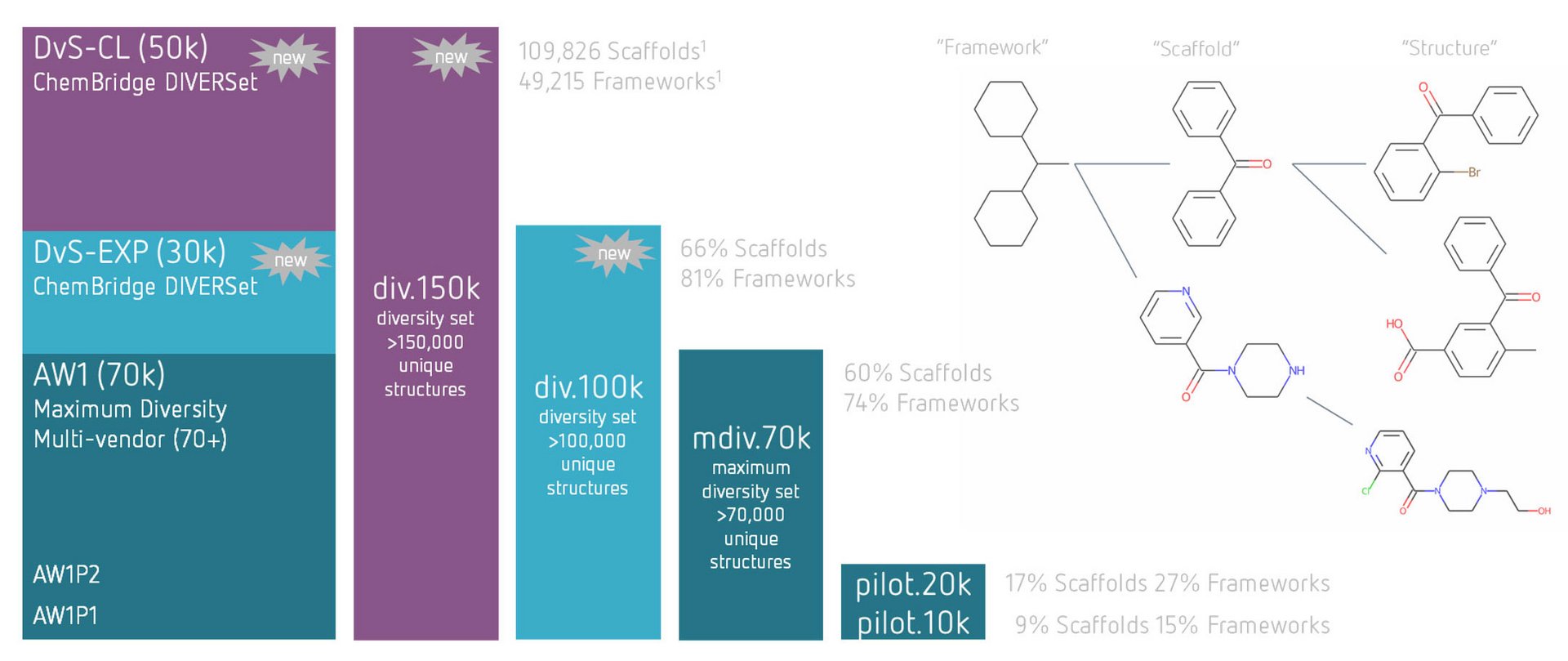
Assay.Works' library of 150,000 small molecule compounds originates from the screening deck of a renowned bio-pharmaceutical enterprise enriched with compounds from a leading commercial supplier. It has been exclusively designed by experienced computational chemists and meticulously curated for optimal lead-likeness and diversity. To ensure the quality of compound samples in the library, molecular mass and purity have been verified by LC/UV/MS analysis during admission.
Assay.Works sets itself apart with its state-of-the-art compound management facility. A cutting-edge acoustic dispensing platform allows to precisely dispense quantities ranging from nanoliters to microliters, resulting in significant resource savings. Sample management operations, including compound registration, screening plate production, and inventory management are backed by Mosaic Sample Management, a leading LIMS solution.
Lipinski Rule-of-5
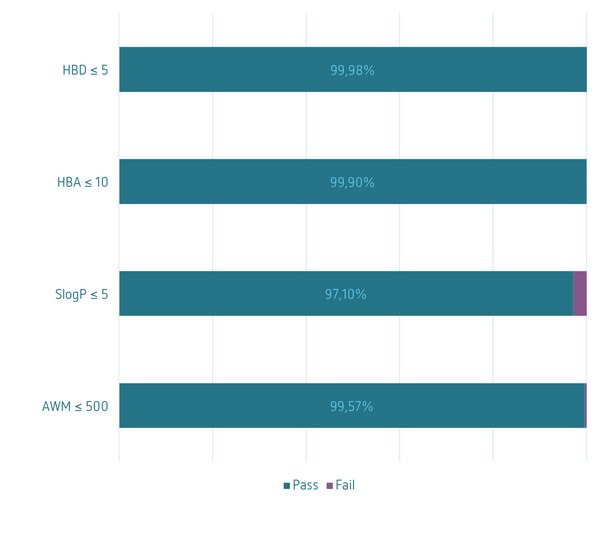
Property Prediction
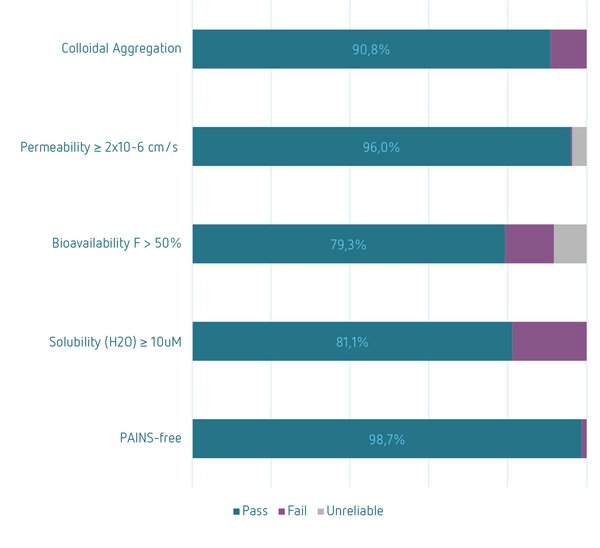
Physico-chemical property profile according to Lipinski’s Rule of 5 (left); Prediction of compound properties and drug-like features: Colloidal Aggregation[i], Permeability[ii], Bioavailability[iii], Solubility[iv], PAINS[v] (right)
Assay.Works offers a modular toolbox of eleven non-redundant sets of 5000 bioactive small molecules, including endogenous metabolites, enzyme inhibitors, receptor ligands, and FDA-approved drugs.
- Broad coverage of drug target classes, therapeutic indications, and research areas
- Structurally diverse, pharmacologically active, and cell permeable
- Screen-ready DMSO stocks, quality-controlled by HPLC and NMR
- Rich set of chemical and biological annotations
Applications
- Target validation
- pathway deconvolution
- combination studies
- Assay validation
- exploratory screening
Pathway | Immunology (IMN) Immunology/Inflammation-related compounds. Targets include CCR, COX, Interleukin Related, IRAK, MyD88, PDE, PD-1/PD-L1, TLR, and more. Includes some compounds related to tumor-immunology. | 292 |
| Epigenetics (EPG) Epigenetics-related compounds targeting HDAC, Histone Demethylase, Histone Acetyltransferase (HAT), DNA Methyltransferase (DNMT), Epigenetic Reader Domain, MicroRNA, etc. | 302 | |
| Neuronal Signaling (NRS) Compounds related to Neuronal Signaling. Targets include 5-HT Receptor, AChE, Adrenergic Receptor, AMPAR, Beta- and Gamma-secretase, Dopamine Receptor, FAAH, Melatonin Receptor, AChR, Opioid Receptor, etc. | 497 | |
| Autophagy (APH) Compounds with biological activity used for autophagy research and associated assays. Targets include Autophagy, LRRK2, ULK, etc. | 427 | |
| Cell Death (CLD) Compounds related to cell cycle, cell survival, DNA damage, and apoptosis. Targets includes CDK, ROCK, Aurora Kinase, ATM/ATR, DNA-PK, DNA/RNA Synthesis, Bcl-2 Family, Caspase, DAPK, IAP, MDM2/p53, PKD, Survivin, etc. | 405 | |
| Metabolism (MET) Metabolism/Protease-related small molecules. Targets include PDE, Cytochrome P450, HMG-CoA Reductase, DPP4, Proteasome, HCV Protease, IDO, Cathepsin, MMP, etc. | 608 | |
Target Class | Transporter & Ion Channels (TIC) Modulators of ion channels and membrane transporters related to targets and pathways not contained in the above sets. | 237 |
| GPCR & G-Protein (GPR) Modulators of GPCR and G-proteins related to targets and pathways not contained in the above sets. | 317 | |
| Kinase Inhibitors (KIN) Protein kinase inhibitors related to pathways not contained in the above sets. | 429 | |
Other | Anti-Infection (AIN) Anti-infective compounds targeting Bacteria, Fungi, Parasite, CMV, HIV, SARS-CoV, Influenza Virus, etc. | 442 |
| Other Bioactives (OBA) Other bioactive compounds related to targets and pathways not contained in the above sets. | 1,095 |
[1] Structures and annotations disclosed under confidentiality agreement upon request.
[2] At the time added to the original source collections. To validate sample integrity, random samples are re-analyzed during the lifetime of stored liquid aliquots.
[i] Small Colloidal Aggregating Molecule (SCAM) prediction based on a consensus model. Fail criteria: 95% consensus that a molecule is a SCAM. Molina C, Ait-Ouarab L, Minoux H. J Chem Inf Model. 2022.
[ii] Caco-2 permeability prediction based on a conditional consensus model. Pass criteria: High to medium permeability, > 2x10-6 cm/s. Falcón-Cano G, Molina C, Cabrera-Pérez MÁ. ADME Prediction with KNIME: An automated prediction platform for in silico assessment of Caco-2 permeability. 2022, Submitted.
[iii] Human oral bioavailability prediction. Pass criteria: F > 50%. Falcón-Cano G, Molina C, Cabrera-Pérez MÁ. J Chem Inf Model. 2020
[iv] Aqueous solubility prediction based on consensus model. Pass criteria: logS >= -5 (S in mol/l). Falcón-Cano G, Molina C, Cabrera-Pérez MÁ. ADMET DMPK. 2020
[v] Pan Assay Interference Compound (PAINS) filter using a published KNIME workflow. Saubern S, Guha R, Baell JB. Mol Inform. 2011